Our latest paper, "Standardized Benchmarking in the Quest for Orthologs", just came out in Nature Methods. This is a brief overview and story behind the paper.
Orthology benchmarking
Orthology, which formalises the concept of "same" genes in different species, is a foundation of genomics. Last year alone, more than 13,000 scientific papers were published with keyword “ortholog”. To satisfy this enormous demand, many methods and resources for ortholog inference have been proposed. Yet because orthology is defined from the evolutionary history of genes, which usually cannot be known with certainty, benchmarking orthology is hard. There are also practical challenges in comparing complex computational pipelines, many of which are not available as standalone software.
Identifier mapping: the bane of bioinformatics
Back in 2009, Adrian Altenhoff and I published a paper on ortholog benchmarking in PLOS Computational Biology. At the time, this was the first benchmark study with phylogeny-based tests. It also investigated an unprecedented number of methods. One of the most challenging aspect of this work—and by far the most tedious—was to compare inferences performed by different methods on only partly overlapping sets of genomes, often with inconsistent identifiers and releases—giving right to the cynics’s view that "bioinformatics is ninety percent identifier mapping…"
Enter the Quest for Orthologs consortium
Around that time, Eric Sonnhammer and Albert Vilella organised the first Quest for Orthologs (QfO) meeting at the beautiful Genome Campus in Hinxton, UK—the first of a series of collaborative meetings. We have published detailed reports on these meetings (2009, 2011, 2013; stay tuned for the 2015 meeting report…).
Out of these interactions, the Quest for Orthologs consortium was born, with the mission to benchmark, improve and standardise orthology predictions through collaboration, the use of shared reference datasets, and evaluation of emerging new methods.
The consortium is open to all interested parties and now includes over 60 researchers from 40 institutions worldwide, with representatives from many resources, such as UniProt, Ensembl, NCBI COGs, PANTHER, Inparanoid, PhylomeDB, EggNOG, PLAZA, OrthoDB and our own OMA resource.
The orthology benchmark service and other contributions of the paper
The consortium is organised in working groups. One of them is the benchmarking working group, in which Adrian and I have been very involved. This new paper presents several key outcome of the benchmarking working group.
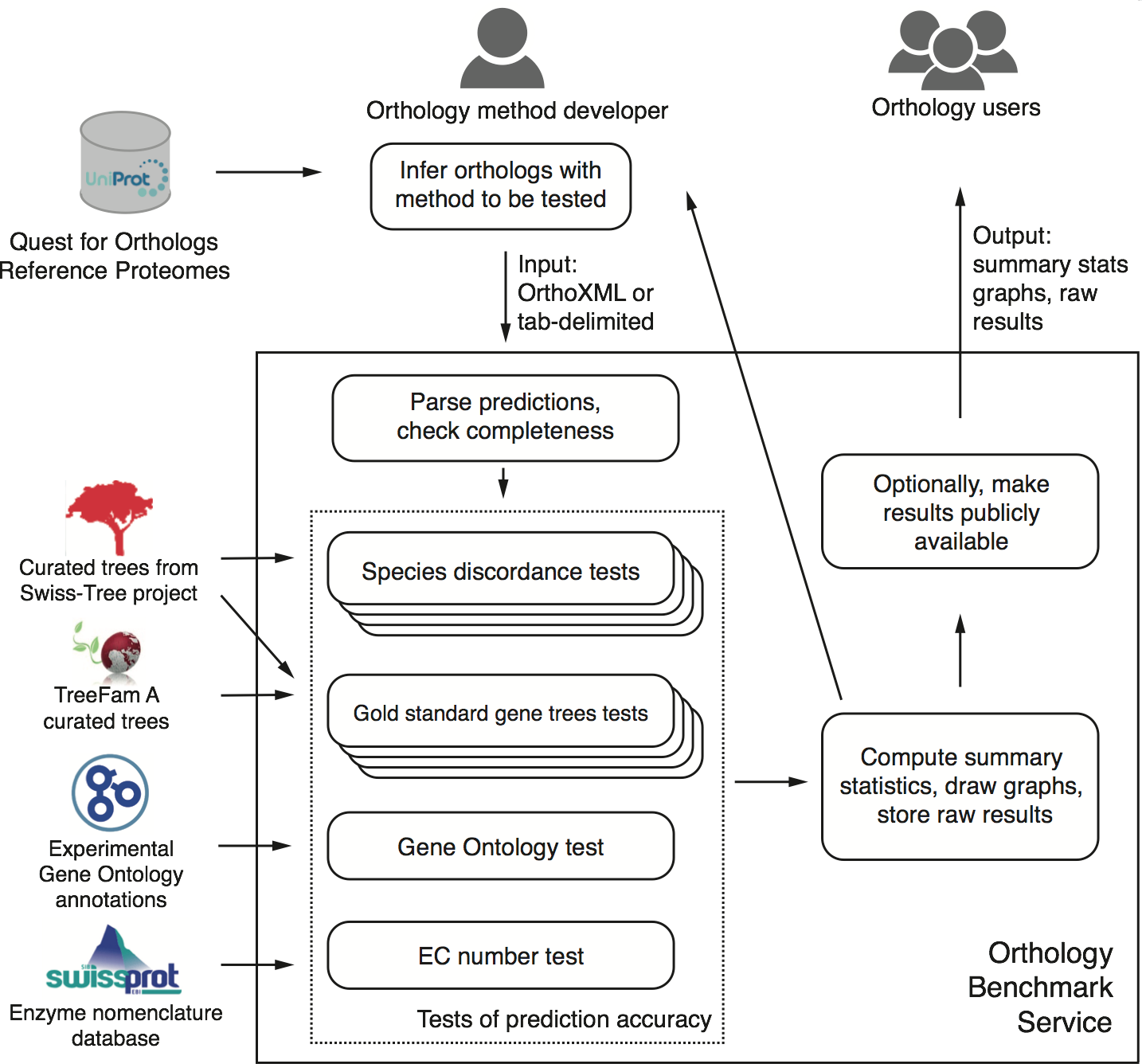
First and foremost, we present a publicly-available, automated, web-based benchmark service. Accessible at http://orthology.benchmarkservice.org, the service lets method developers evaluate predictions performed on the 2011 QfO reference proteome set of 66 species. Within a few hours after submitting their predictions, they obtain detailed feedback on the performance of their method on various benchmarks compared with other methods. Optionally, they can make the results publicly available.
Conceptual overview of the benchmark service (Fig 1 of the paper; click to enlarge)
Second, we discuss the performance of 14 orthology methods on a battery of 20 different tests on a common dataset across all of life.
Third, one of the benchmark, the generalised species discordance test, is new and provides a way for testing pairwise orthology based on trusted species trees of arbitrary size and shape.
Implications
For developers of orthology prediction methods, this work sets minimum standards in orthology benchmarking. Methodological innovations should be reflected in competitive performance in at least a subset of the benchmarks (we recognise that different applications entail different trade-offs). Publication of new or update methods in journals should ideally be accompanied by publication of the associated results in the orthology benchmark service.
For end-users of orthology predictions, the benchmark service provides the most comprehensive survey of methods to date. And because it can process new submissions automatically and continuously, it holds the promise of remaining current and relevant over time. The benchmark service thus enables users to gauge the quality of the orthology calls upon which they depend, and to identify the methods most appropriate to the problem at hand.
References
Altenhoff, A., Boeckmann, B., Capella-Gutierrez, S., Dalquen, D., DeLuca, T., Forslund, K., Huerta-Cepas, J., Linard, B., Pereira, C., Pryszcz, L., Schreiber, F., da Silva, A., Szklarczyk, D., Train, C., Bork, P., Lecompte, O., von Mering, C., Xenarios, I., Sjölander, K., Jensen, L., Martin, M., Muffato, M., Altenhoff, A., Boeckmann, B., Capella-Gutierrez, S., DeLuca, T., Forslund, K., Huerta-Cepas, J., Linard, B., Pereira, C., Pryszcz, L., Schreiber, F., da Silva, A., Szklarczyk, D., Train, C., Lecompte, O., Xenarios, I., Sjölander, K., Martin, M., Muffato, M., Quest for Orthologs consortium, Gabaldón, T., Lewis, S., Thomas, P., Sonnhammer, E., Dessimoz, C., Gabaldón, T., Lewis, S., Thomas, P., Sonnhammer, E., & Dessimoz, C. (2016). Standardized benchmarking in the quest for orthologs Nature Methods DOI: 10.1038/nmeth.3830
Altenhoff, A., & Dessimoz, C. (2009). Phylogenetic and Functional Assessment of Orthologs Inference Projects and Methods PLoS Computational Biology, 5 (1) DOI: 10.1371/journal.pcbi.1000262
Gabaldón, T., Dessimoz, C., Huxley-Jones, J., Vilella, A., Sonnhammer, E., & Lewis, S. (2009). Joining forces in the quest for orthologs Genome Biology, 10 (9) DOI: 10.1186/gb-2009-10-9-403
Dessimoz, C., Gabaldon, T., Roos, D., Sonnhammer, E., Herrero, J., & Quest for Orthologs Consortium (2012). Toward community standards in the quest for orthologs Bioinformatics, 28 (6), 900-904 DOI: 10.1093/bioinformatics/bts050
Sonnhammer, E., Gabaldon, T., Sousa da Silva, A., Martin, M., Robinson-Rechavi, M., Boeckmann, B., Thomas, P., Dessimoz, C., & Quest for Orthologs Consortium. (2014). Big data and other challenges in the quest for orthologs Bioinformatics, 30 (21), 2993-2998 DOI: 10.1093/bioinformatics/btu492